My Projects
Explore my work and the projects I'm working on
Epigenetics and Aging
With funding from the NSERC Undergraduate Student Research Award (USRA) and inspiration from the book lifespan, I developed a joint project with Dr. Chippindale and Dr. Aristizabal to explore how epigenetic remodeling influences aging in Drosophila. My work in the Aristizabal lab consisted of protein extractions, SDS-PAGE, immunostaining and western blotting to investigate histone modifications, while in the Chippindale lab, I carried out phenotypic analyses, including sex classification and assays measuring fitness, competition, desiccation resistance, development time, and longevity.
I published an abstract on this research in the Inquiry@Queen’s Research Conference Proceedings here
Synthetic Biology in Space Exploration
My team and I at QAB are embarking on a project to tackle one of the key challenges in space exploration and agriculture: perchlorate contamination. Specifically, we are focused on investigating the catalytic activity of chlorite dismutase variants and modeling de novo mutations to enhance perchlorate detoxification into oxygen and chlorine. These harmful salts could be crucial to the Mars colonization effort, and harnessing their properties could bring us one step closer to becoming a multiplanetary species.
Project Posters
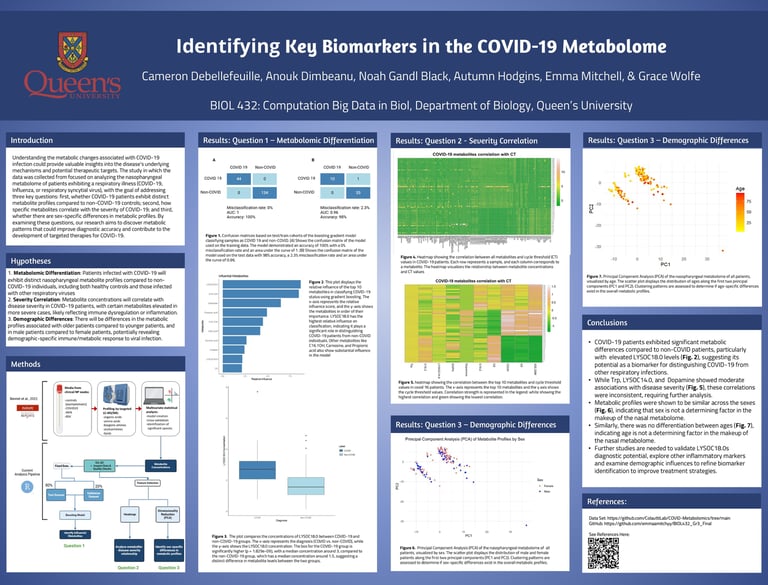
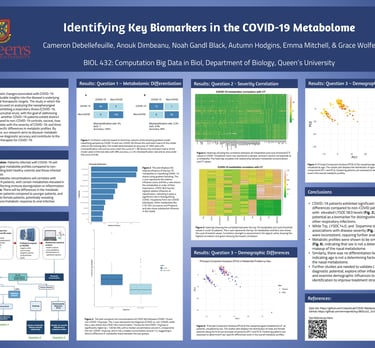
Predicting COVID-19 Status Using ML and Nasal Metabolome Data
In this project, I used gradient boosting, a supervised ensemble machine learning technique, to analyze nasal metabolome data and classify COVID-19 status. The model achieved 98% accuracy on test data, with LYSOC18.0 emerging as the most influential metabolite, showing significantly higher concentrations in COVID-19 patients (p = 1.83e-09). These findings suggest promise for non-invasive, metabolite-based diagnostics to support rapid and accurate COVID-19 detection.
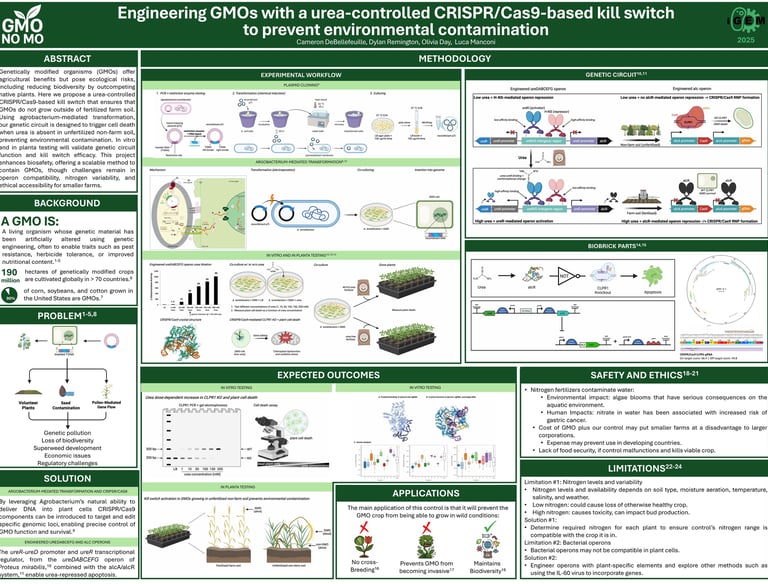
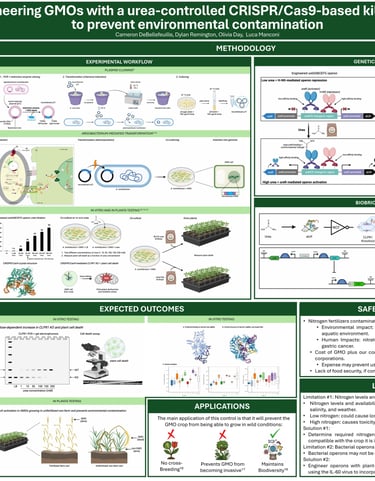
Designing a urea-controlled kill switch for agricultural biocontrol
In this project, I designed a plasmid for assembling our genetic construct, which incorporated the AlcR/AlcA system along with the CRISPR-Cas9 system (spCas9 and sgRNA). I used Benchling to design sgRNAs targeting exon 2 of the CLPR1 gene, aiming to induce apoptosis through gene knockout. To evaluate the effectiveness of the system, I conducted computational modeling of the sgRNA and spCas9 interactions using HADDOCK, which revealed favorable binding and confirmed the potential for successful gene editing.
Engineering Immune Cells to Treat Cystic Fibrosis (Queen's iGEM)
As a part of the Queen’s iGEM synthetic biology team, I’m contributing to the design and engineering of macrophages that can degrade Pseudomonas aeruginosa biofilms, an approach relevant to treating cystic fibrosis. This work involves molecular cloning using HiFi DNA assembly and PCR amplification, as well as designing plasmid constructs for expression in mammalian cells, which we validate through gel electrophoresis and sequencing. Alongside lab work, I co-host The Breathing Room, a podcast exploring the intersection of cystic fibrosis science and ethics, and produced Genes to Genius, a video explaining synthetic biology concepts at five different levels of complexity.
🎧 The Breathing Room: Spotify Podcast
🎬 Genes to Genius: YouTube video
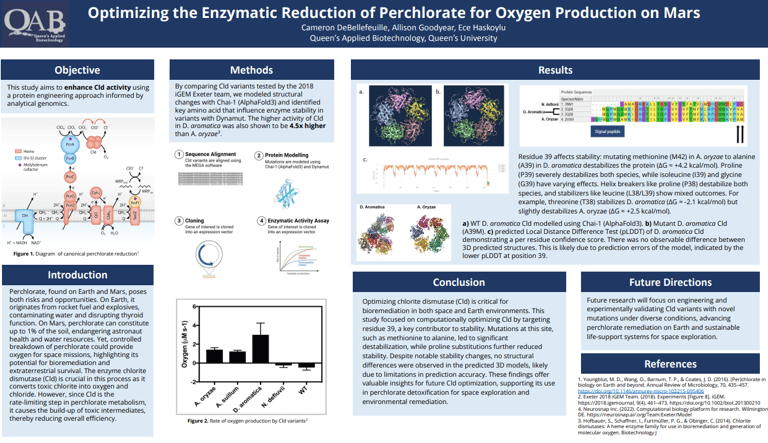
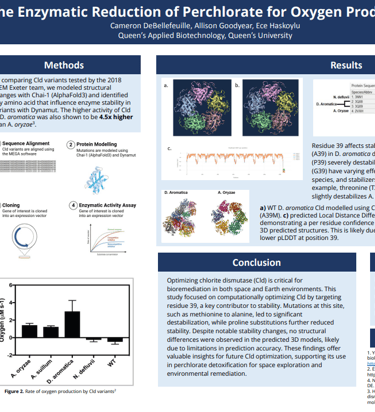
Detoxifying Mars With Synthetic Biology
Our project explored the perchlorate reducing system in naturally occurring perchlorate reducing bacteria (PRB) with the goal of optimizing it for more efficient perchlorate remediation. We focused on chlorite dismutase, the rate limiting enzyme in the pathway, conducting computational analysis with AlphaFold 3 and DynaMut to predict how specific amino acid mutations could enhance catalytic efficiency. Additionally, we designed a plasmid encoding the full perchlorate reducing system for transformation into a bacterial host, enabling targeted bioremediation capabilities.
Achievements
Dean's Honour List-Distinction
Awarded to students in the top 3% of all students within their degree program
NSERC Undergraduate Summer Research Award
$9,800 awarded based on academic excellence and research potential
$4,000 awarded to students with a high school average of 95%+
Queen's Principal's Scholarship
Prize-winning agar art created by Mason Wilson and I in our Fundamentals of Microbiology class.
“Nothing in life is to be feared, it is only to be understood. Now is the time to understand more, so that we may fear less.”
- Marie Curie
Subscribe to my monthly Newsletter


I share what I learn each month in genetics, synthetic biology, and astrobiology as well as my professional and project management insights that I've learned through overcoming challenges in my projects and career